Structure-aware generative molecular design: from 2D or 3D?
Background
For this reason, 3D-GMD (algorithms that build the ligand in 3D) and structure-explicit generative molecular design (algorithms that utilise a representation of protein structure) are a large focus of research currently. However, you can also achieve structure-guided GMD by using structure-aware scoring functions to guide GMD that is typically done in 2D. Last year I wrote a mini-review comparing these differences (Thomas M, et al., 2023) but, it is already a little out of date and doesn't include, for example, diffusion models for 3D-GMD.
What is current state of 3D-GMD?
PoseCheck (Harris C et al. arXiv, Aug 2023): “The exceedingly high strain energy [~1,200 kcal/mol] values observed in this scenario should be approached with considerable prudence. For comparison, the combustion of TNT releases approximately 815 kcal/mol.”
Does 3D dominate? (Zheng K et al. arXiv, Jun 2024): “Also, representing target molecules in 3D format does not significantly improve both the molecular quality and binding affinity.”
DrugPose (Jocys Z et al. Digital Discovery, Jun 2024): “The evaluation of different [3D] models reveals that the current generative models have limitations in generating molecules with the desired binding mode.”
GenBench3D (Baillif B et al. ChemRxiv, Jul 2024): “We benchmarked six structure-based [3D] molecular generative models and showed that they generated mostly molecules with invalid geometries.”
CBGBench (Lin H et al. arXiv, Jul 2024): “it is worth noting that DIFFBP, FLAG, and POCKET2MOL can possibly generate molecules with small atom numbers and high LBE [ligand binding efficiency].”
POKMOL3D (Liu H et al. ChemRxiv, Aug 2024): “Overall, the performance of 3D generative models on large scale of pockets is still far from satisfactory, and polishing of network architecture is needed to improve the learning of ligand-protein interaction and generalizability of current 3D generative models to enable their application on wide range of protein pockets.”
Well I wouldn't say they are all glowing recommendations, it seems the most positive conclusion is that some models "can possibly generate molecules ... with high LBE". However, only the comparisons by Zheng et al. and Liu H et al. included a comparison to structure-implicit GMD in 2D, moreover, only the comparison by Liu H et al. also evaluated the type of chemistry generated beyond geometry and basic measures (like QED, which in my experience can be hacked). Therefore, I guide the interested reader to the comparison by Liu H et al. Nonetheless, I wanted to try some methods myself.
Experiment
In the interest of a breadth, I have selected six orthogonal GMD algorithms, three structure-explicit models based on the best performing algorithms in the above benchmarks, and three structure-implicit GMD algorithms that should prove good baselines.Structure-explicit GMD algorithms
- [3D] Lingo3DMol (Feng W, et al.): An auto-regressive language model that generates local coordinates of atoms conditioned on the protein pocket.
- [3D] Pocket2Mol (Peng X, et al.): An auto-regressive graph neural network that predicts focal atom, new atom position, bond and type directly in the pocket.
- [3D] TargetDiff (Guan J, et al.): A diffusion model that de-noises atom positions conditioned on the protein pocket.
- [2D] AHC (Thomas M et al., 2022): An auto-regressive GRU-based chemical language model pretrained on ChEMBL, with efficient reinforcement learning to optimise an objective.
- [2D] AutoGrow 4 (Spiegel JO, et al.): Heuristic molecule building optimised by a genetic algorithm over chemistry-aware operations including crossover by substructure overlap, and mutations by chemical reactions.
- [3D] 3D-MCTS (Du H, et al.): Heuristic molecule building in the pocket by step-wise addition of fragments optimised by a Monte-Carlo Tree Search.
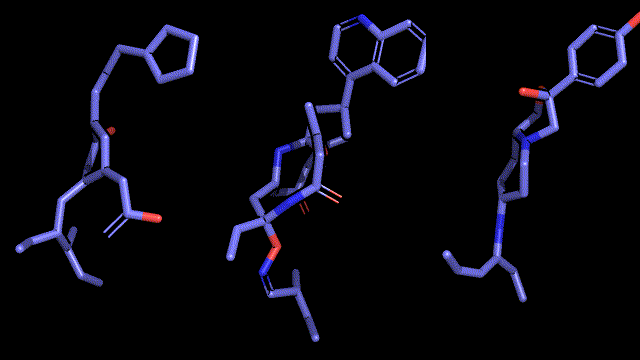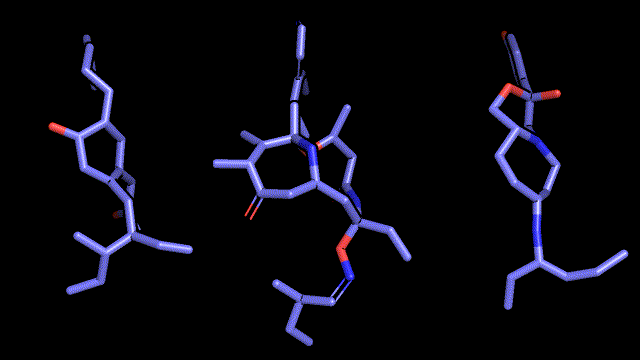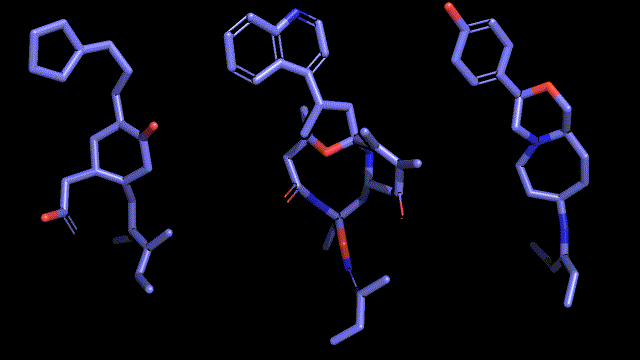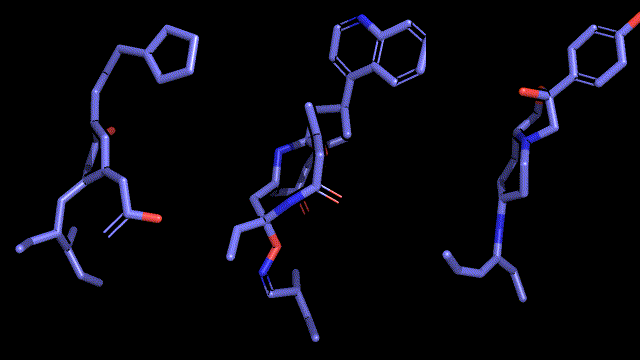
Qualitative analysis (A picture says a thousand words)
 |
| Pocket2Mol |
 |
| 3D-MCTS |
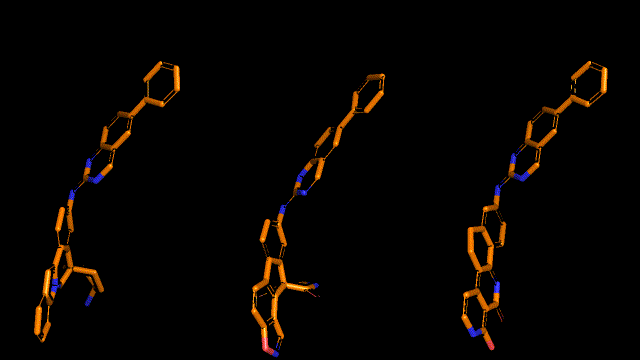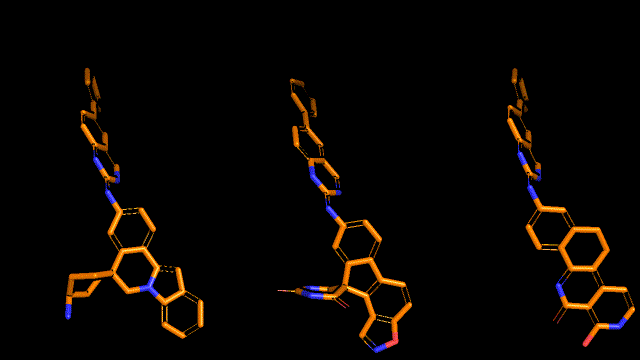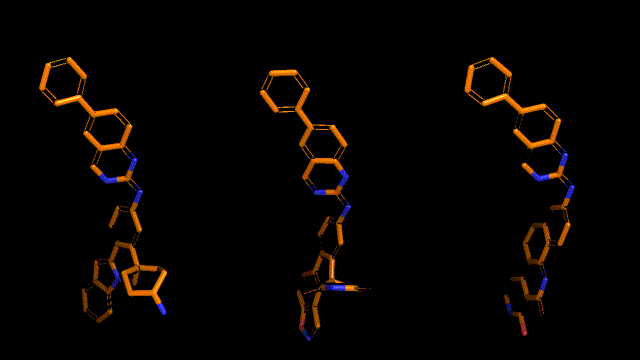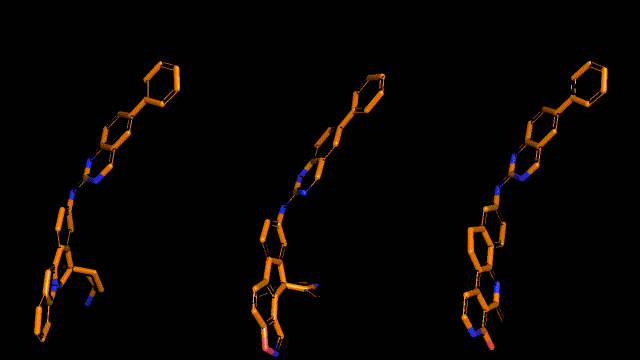 |
| AHC |
Quantitative analysis I (Geometry comparison)
- The number of steric clashes i.e., instances two neutral atoms are closer than their combined van de Vaals radii which would be energetically unfavourable.
- The ligand strain energy i.e., the energy difference between the generated pose and a conformationally relaxed ligand without the protein, calculated using the Universal Force Field (UFF).
- The interaction similarity i.e., the Tanimoto similarity of the protein-ligand interactions compared to the reference ligand, in this case Risperidone.
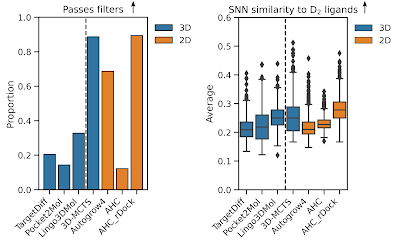
Quantitative analysis II (Chemical quality comparison)
- Proportion of unique scaffolds.
- Proportion of molecules that pass chemistry filters. This includes medicinal chemistry filters and PAINS filters that discard reactive, problematic, or promiscuous substructures as well as ensuring molecules are in a sensible property space (e.g., molecular weight, LogP range, and number of rotatable bonds).
- Average number of outlier ECFP fingerprint bits that aren't present in a reference dataset (here ChEMBL). This is a proxy for idiosyncratic atomic environments.
- Average single-nearest neighbour similarity to known D2 ligands.
Most striking is that only 20-30% of molecules from structure-explicit 3D-GMD pass the molecular quality filters or don't particularly generate a diverse range of scaffolds (aside from TargetDiff). However, there isn't much overall difference when we look at similarity to known D2 ligands which suggests none of the algorithms are generating molecules in the 'retrospectively correct' chemical space.
Disappointingly, AHC also performs poorly in generating molecules that pass molecular quality filters. However, if we look at the docking scores that were optimised we can see that AHC also learns to optimise the Vina score much more than the alternative structure-implicit algorithms. I believe this is a combination of the broad generative domain of AHC and learning efficiency allowing it to access more Vina favourable chemical space than the other more chemically restrictive generative algorithms (that conduct molecule building via hard-coded rules). More Vina favourable chemical space means larger, greasier molecules that provide good scores. For these reasons, I've never tested a system where Vina is a useful scoring function to guide GMD - perhaps a topic for another post.
For the sake of curiosity, I repeated the experiment but using rDock instead, which utilises a broader balance of physics-based interaction terms in its scoring function. The new analysis below shows an increase from around 10% of molecules passing molecular quality filters to around 90%. Moreover, this approach now generates the most similar molecules to known D2 ligands on average.
Concluding remarks
References
Baillif, B., Cole, J., McCabe, P., & Bender, A. (2024). Benchmarking structure-based three-dimensional molecular generative models using GenBench3D: ligand conformation quality matters. arXiv preprint arXiv:2407.04424.
Cremer, J., Le, T., Noé, F., Clevert, D. A., & Schütt, K. T. (2024). PILOT: Equivariant diffusion for pocket conditioned de novo ligand generation with multi-objective guidance via importance sampling. arXiv preprint arXiv:2405.14925.
Du, H., Jiang, D., Zhang, O., Wu, Z., Gao, J., Zhang, X., ... & Hou, T. (2023). A flexible data-free framework for structure-based de novo drug design with reinforcement learning. Chemical Science, 14(43), 12166-12181.
Feng, W., Wang, L., Lin, Z., Zhu, Y., Wang, H., Dong, J., ... & Zhou, W. (2024). Generation of 3D molecules in pockets via a language model. Nature Machine Intelligence, 6(1), 62-73.
Guan, J., Qian, W. W., Peng, X., Su, Y., Peng, J., & Ma, J. (2023). 3d equivariant diffusion for target-aware molecule generation and affinity prediction. arXiv preprint arXiv:2303.03543.
Harris, C., Didi, K., Jamasb, A., Joshi, C., Mathis, S., Lio, P., & Blundell, T. (2023). Posecheck: Generative models for 3D structure-based drug design produce unrealistic poses. In NeurIPS 2023 Generative AI and Biology (GenBio) Workshop.
Jocys, Z., Grundy, J., & Farrahi, K. (2024). DrugPose: benchmarking 3D generative methods for early-stage drug discovery. Digital Discovery.
Lin, H., Zhao, G., Zhang, O., Huang, Y., Wu, L., Liu, Z., ... & Li, S. Z. (2024). CBGBench: Fill in the Blank of Protein-Molecule Complex Binding Graph. arXiv preprint arXiv:2406.10840.
Liu, H., Niu, Z., Qin, Y., Xu, M., Wu, J., Xiao, X., ... & Chen, H. (2024). How good are current pocket based 3D generative models?: The benchmark set and evaluation on protein pocket based 3D molecular generative models. chemRxiv preprint 10.26434/chemrxiv-2024-2qgpb.
Peng, X., Luo, S., Guan, J., Xie, Q., Peng, J., & Ma, J. (2022, June). Pocket2mol: Efficient molecular sampling based on 3d protein pockets. In International Conference on Machine Learning (pp. 17644-17655). PMLR.
Shen, T., Pandey, M., & Ester, M. TacoGFN: Target Conditioned GFlowNet for Drug Design. In NeurIPS 2023 Generative AI and Biology (GenBio) Workshop.
Spiegel, J. O., & Durrant, J. D. (2020). AutoGrow4: an open-source genetic algorithm for de novo drug design and lead optimization. Journal of Cheminformatics, 12, 1-16.
Thomas, M., M., Bender, A., & de Graaf, C. (2023). Integrating structure-based approaches in generative molecular design. Current Opinion in Structural Biology, 79, 102559.
Thomas, M., O’Boyle, N. M., Bender, A., & De Graaf, C. (2022). Augmented Hill-Climb increases reinforcement learning efficiency for language-based de novo molecule generation. Journal of Cheminformatics, 14(1), 68.
Thomas, M., O’Boyle, N. M., Bender, A., & De Graaf, C. (2024). MolScore: a scoring, evaluation and benchmarking framework for generative models in de novo drug design. Journal of Cheminformatics, 16(1), 64.
Xu, M., Ran, T., & Chen, H. (2021). De novo molecule design through the molecular generative model conditioned by 3D information of protein binding sites. Journal of Chemical Information and Modeling, 61(7), 3240-3254.
Zheng, K., Lu, Y., Zhang, Z., Wan, Z., Ma, Y., Zitnik, M., & Fu, T. (2024). Structure-based Drug Design Benchmark: Do 3D Methods Really Dominate?. arXiv preprint arXiv:2406.03403



.png)
.png)
.png)




Comments
Post a Comment